Introduction
Atopic dermatitis is the most common chronic inflammatory skin disease of early childhood, with high heritability [1]. The worldwide cumulative prevalence of atopic dermatitis is approximately 10–20% in this age group and it has been steadily increasing over past decades [2–4]. A multifactorial background for atopic dermatitis has been suggested, with genetic as well as environmental factors influencing disease development [5]. There is substantial evidence from family and twin studies in support of a strong genetic contribution but our understanding of the genetic predisposition to atopic dermatitis remains limited [6]. A number of association studies and GWASs have reported that mutations in filaggrin (FLG) are the most significant risk factor for atopic dermatitis [7, 8]. Apart from FLG, recent GWASs and immunochip analysis have identified a total of 19 additional contributory loci of atopic dermatitis with genome-wide significance [9]. The first GWAS of atopic dermatitis identified a susceptible variant on chromosome 11q13.5, located in an intergenic region between uncharacterised protein coding gene C11orf30 and the gene encoding the leucine-rich type I membrane protein (LRRC32) [10]. This common variant, rs7927894[T], was shown to be associated with atopic eczema in German, British, and Irish populations, but the association was not replicated in the Japanese population [10–13]. To gain a better understanding of the role of the genetic variation in atopic dermatitis, it is important to validate the association of previously reported loci in different populations.
Aim
In the current study, we aimed to assess the importance of this common variant in the susceptibility to atopic dermatitis in children younger than 2 years of age and whether this possible association is independent of the well-described FLG risk alleles. Moreover, we evaluated the association of this novel risk variant with the severity of the atopic dermatitis and disease-related phenotypes.
Material and methods
Study population
A total of 188 unrelated children (107 males) less than 2 years old at the time of recruitment were enrolled, namely 103 patients with atopic dermatitis (mean age: 13.2 ±6.7 months) and 85 healthy control subjects (mean age: 15.3 ±5.6 months). All study participants were of Caucasian ethnicity. The study subjects were recruited from patients who visited the Outpatient Clinic for Children at the Wroclaw Medical University Hospital and from the general population, as described below. The healthy control subjects were recruited from the general population through community-based approaches. We distributed flyers at local nurseries, child and family doctor’s surgeries, health fairs. Interested parents were instructed to contact the research coordinator via phone. All participants, cases and controls, were selected using a detailed questionnaire that included questions regarding the overall health status, symptoms of atopic dermatitis and other allergic diseases, sociodemographic information, and family history of allergic diseases. The subjects with atopic dermatitis were examined and diagnosed according to the criteria established by Hanifin and Rajka [14]. The mean age at disease onset was 4.6 ±3.5 months. Atopic dermatitis severity was assessed using the SCORing Atopic Dermatitis index (SCORAD) and the patients were divided into mild (< 25 points), moderate (25–50 points), or severe (> 50 points) disease groups. Subjects with atopic dermatitis were divided into allergic and non-allergic on the basis of the positive specific IgE level against at least one of the allergens tested. The control group included healthy children and met the following criteria: absence of symptoms of atopic dermatitis and asthma or other allergic diseases, absence of allergic sensitisation and negative family history of allergic diseases.
Serum measurements for total and specific IgE levels were performed on all recruited subjects, including IgE specific to the most popular 10 inhalant and 10 food allergens. The concentration of total serum IgE was measured using the commercially available kit IMMULITE 2000 Total IgE (Diagnostic Products Corporation (DPC), USA). The levels of specific IgE were determined using a standard enzyme immunoassay (Polycheck, BIOCHECK, Germany). Allergic sensitisation was defined as the presence of a specific IgE to at least one of the tested allergens of ≥ 0.7 kU/l (Class II).
The study was approved by the ethics committee and informed written consent, including consent to genetic studies, was obtained from all of the subjects before testing.
Genotyping
Samples of the 188 subjects were genotyped for the rs7927894 SNP on chromosome 11 and for the four common FLG mutations R501X, 2282del4, R2447X, and S3247X. Children with the FLG mutation in any of these positions were classified as FLG loss-of-function mutation cases. Genomic DNA was obtained from EDTA whole blood samples using the QIAamp DNA Blood Mini Kit (QIAGEN GmbH, Germany). All mutations were determined using a LightSNiP assay (TIB Molbiol, Berlin, Germany). PCR was performed in a final volume of 10 µl containing 1 µl of DNA at concentration 15–60 ng/µl, 0.5 µl of reagent mix containing specific primers and SimpleProbe® probes at optimised concentration, 0.8 µl of MgCl2 and 1 µl of LightCycler® FastStart DNA Master HybProbe (Roche Applied Science, Mannheim, Germany). Reactions were performed on a LightCycler 1.5 platform (Roche Applied Science, Mannheim, Germany). For quality control of genotyping procedures, positive controls of each genotype, as well as negative controls, were included in each reaction.
Statistical analysis
The Hardy-Weinberg equilibrium was tested using the χ2 goodness-of-fit test to compare observed genotype frequencies with expected frequencies among the controls. Differences in genotype frequencies or demographic characteristics between case and control groups were evaluated using the χ2 test or Fisher exact test, as appropriate. The associations of genotypes or alleles with patient groups versus control subjects were determined by computing the odds ratio (OR), its 95% confidence interval (95% CI), and p-values using logistic regression analysis for crude ORs and adjusted ORs when adjusting for family history of atopy. Statistical significance was set at p-values < 0.05. Three different genetic models were tested separately when comparing genotypes and atopic dermatitis in this study. To study a co-dominant model, the three genotype groups were analysed separately, using wild-type homozygotes as a reference group. To test a dominant model, wild-type homozygotes were compared with heterozygotes and homozygotes for minor alleles. In the multiplicative model genotypes were coded as a 3-level variable for the number of minor alleles. χ2 test or Fisher exact test was used to determine the combined effect of genotype pairs. Gene-gene interaction was investigated using logistic regression models for atopic dermatitis with interaction terms (SPSS). To establish whether an interaction between the two risk factors A (rs7927894) and B (FLG mutations) existed, the relative excess risk due to interaction (RERI), the proportion attributable to interaction (AP), the synergy index (S) and the ratio of ORs were calculated, as recommended by Knoll et al. [15, 16]. Interaction was defined as departure from the additive or multiplicative model. On the additive scale a RERI and an AP > 0 or < 0 indicate a superadditive or subadditive effect, respectively, an S > 1 or < 1 means a superadditive or subadditive effect, respectively. On the multiplicative scale the ratio of ORs > 1 or < 1 indicates a positive or negative interaction, respectively. Statistical analysis was carried out using the program package Statistica Version 9.0 (StatSoft, Inc., Tulsa, OK, USA) and the SPSS Statistics software package Version 11.1 (SPSS Inc., Chicago, IL, USA).
Results
Baseline characteristics of patients with atopic dermatitis and controls are given in Table 1. There were no significant differences between the cases and the controls for age and gender. The allele and genotype distributions for the rs7927894 SNP on chromosome 11q13.5 and the FLG mutations R501X, 2282del4, R2447X, and S3247X in the cases and controls are shown in Table 2.
Table 1
Characteristics of the study group
Table 2
Genotype and allele frequencies of rs7927894 and FLG loss-of-function mutation in the studied group
| Genotype | Control n (%) | Atopic dermatitis | Allergic atopic dermatitis | Non-allergic atopic dermatitis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | P-value | aOR*(95% CI) | n (%) | P-value | aOR*(95% CI) | n (%) | P-value | aOR*(95% CI) | ||
| Chromosome 11q13.5 rs 7927894: | ||||||||||
| CC | 32 (37) | 22 (21) | 0.028 | 1.00 (Reference) | 12 (22) | 0.026 | 1.00 (Reference) | 10 (21) | 0.134 | 1.00 (Reference) |
| CT | 40 (47) | 54 (52) | 2.02
(1.00–4.07) | 25 (45) | 1.67 (0.67–4.16) | 29 (60) | 2.32 (0.97–5.53) | |||
| TT | 13 (15) | 27 (26) | 2.71
(1.09–6.71) | 18 (32) | 2.98
(1.02–8.68) | 9 (19) | 2.21 (0.63–7.76) | |||
| CT + TT | 53 (62) | 81 (78) | 0.018 | 2.21
(1.14–4.28) | 43 (78) | 0.049 | 2.16
(0.98–5.06) | 38 (79) | 0.010 | 2.70
(1.09–6.71) |
| T | 66 (39) | 108 (52) | 0.009 | 1.73
(1.12–2.68) | 61 (55) | 0.007 | 1.96
(1.17–3.28) | 47 (49) | 0.122 | 1.51 (0.88–2.58) |
| Combined FLG genotype: | ||||||||||
| Normal | 83 (98) | 89 (86.5) | 0.007 | 1.00 (Reference) | 52 (81) | 0.001 | 1.00 (Reference) | 37 (86) | 0.589 | 1.00 (Reference) |
| Null | 2 (2) | 14 (13.5) | 6.52
(1.35–42.9) | 12 (19) | 9.57
(1.91–64.7) | 2 (14) | 2.24 (0.21–23.4) | |||
The rs7927894 SNP showed a significant association with atopic dermatitis (p = 0.028). In logistic regression analysis the rs7927894 TT homozygote and CT heterozygote were associated with a significantly increased risk of atopic dermatitis compared with the wild-type homozygote. The genotype model showed an OR of 2.02 for heterozygote individuals and 2.71 for homozygotes. The OR associated with the rs7927894 SNP was estimated at 1.73, using the allele model (Table 2). Analysis of the combination of all 4 tested FLG mutations also revealed a strong and significant association with susceptibility to atopic dermatitis (p = 0.007). Patients carrying at least one of these four FLG mutations have an approximately 6-times greater chance of having atopic dermatitis than individuals who do not carry any of these variants. Furthermore, the multivariate analysis of the genotypes adjusted for family history of atopy confirmed these associations (Table 2). There were no significant associations between atopic dermatitis severity and either the rs7927894 SNP or the combined FLG variants. The mild and moderate atopic dermatitis groups did not differ in terms of frequency of rs7927894 SNP (p = 0.884; χ2 = 0.247) and FLG mutations (p = 0,619; χ2 = 0.252) (Table 3).
Table 3
Associations between rs7927894 and the combined FLG genotype and atopic dermatitis severity
| Genotype | Mild atopic dermatitisn (%) | Moderate atopic dermatitisn (%) | P-value | aOR* (95% CI) |
|---|---|---|---|---|
| Chromosome 11q13.5 rs 7927894: | ||||
| CC | 16 (20.2) | 6 (25) | 1.00 (Reference) | |
| CT | 42 (53.2) | 12 (50) | 0.767 | 1.31 (0.36–4.66) |
| TT | 21 (26.6) | 6 (25) | 0.762 | 1.31 (0.29–5.82) |
| CT + TT | 63 (79.7) | 18 (75) | 0.762 | 1.31 (0.23–2.56) |
| T | 84 (53.2) | 24 (50) | 0.743 | 1.13 (0.56–2.27) |
| Combined FLG genotype: | ||||
| Normal | 69 (87.4) | 20 (83.3) | 0.619 | 1.38 (0.32–5.54) |
| Null | 10 (12.6) | 4 (16.7) | ||
When the allergic and non-allergic atopic dermatitis groups were analysed separately, both groups had the rs7927894 allele significantly more frequently than the control group. The rs7927894[T] allele increased the risk of atopic dermatitis in both groups, however in non-allergic subjects the significant association was only detected under the dominant model. The non-allergic and allergic atopic dermatitis groups did not differ in terms of frequency of this SNP (p = 0.221). Concerning FLG mutations, carrying at least one of FLG null mutations was a significant risk factor for atopic dermatitis only in subjects who were allergic, whereas no significant association could be observed in the non-allergic group (Table 2).
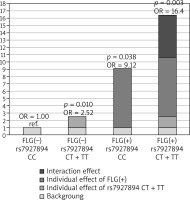
Next, we investigated whether the association of rs7927894 SNP with atopic dermatitis is affected by the strongly-significant FLG null mutations status. Additional analyses performed after stratification for the presence or absence of the combined FLG mutations indicated an association of the risk allele with atopic dermatitis (OR = 1.78; 95% CI: 1.35–2.80) in all tested models but only in the group without FLG mutations (Table 4). We therefore included the combined FLG genotype as a second predictor in a logistic regression model to determine whether the FLG mutations confound rs7927894 SNP and atopic dermatitis association. After adjusting for the presence of FLG mutations, the rs7927894 SNP still shows a statistically significant effect (p = 0.008) with an OR of 2.48 (95% CI: 1.26–4.86). Finally, we investigated whether an interaction between rs7927894 SNP and the four most common FLG mutations influenced the atopic dermatitis risk. Compared with the reference group carrying neither genetic risk factor, the presence of at least one rs7927894[T] allele or at least one loss-of-function FLG mutations significantly increased the atopic dermatitis risk and children with both risk factors were at highest risk (Table 5). The RERI, AP and synergy index indicated positive interaction on the additive scale, meaning that the risk conferred by the combination of both risk factors was significantly higher than the sum of the independent effects. The RERI was 5.77 meaning that OR for atopic dermatitis in children with genetic both risk factors is 5.77 more than if there were no interaction between rs7927894 SNP and the four most common FLG mutations. Figure 1 shows this graphically. The measure of interaction on a multiplicative scale, the ratio of ORs, was 0.71, meaning that there was negative interaction on a multiplicative scale because the combined effect was smaller than the product of the individual effect (Table 5). Our results suggest that there is some evidence of a gene-gene interaction on additive scale, although when modelling interaction, the interaction coefficient was not significant for both genotype combinations (p for interaction = 0.828), which may be because of the low sample size of our study and insufficient statistical power.
Table 4
Atopic dermatitis risk dependent on FLG mutation status
Table 5
Interaction between rs7927894 and the FLG mutations in atopic dermatitis
Discussion
Atopic dermatitis is a common inflammatory disease caused by a combination of genetic and environmental factors [5]. The detection of filaggrin as a major gene implicated in atopic dermatitis in 2006 was an important breakthrough in our understanding of the role of genetic variation in atopic dermatitis [7, 17]. Subsequent studies have shown loss-of-function mutations in the filaggrin gene to be the most significant and widely replicated genetic risk factor reported to date [18–20]. Our former study and data presented here also provided highly significant replication of the previously-reported association of the 4 nonsense FLG mutations with atopic dermatitis [21, 22]. In addition, we have demonstrated that FLG alleles increased the risk of allergic sensitisation, confirming previous results [19, 20, 23, 24].
The strength of the association and the high risk conferred by the filaggrin loss-of-function mutations are remarkable, however FLG null mutations can only explain a small portion of the total burden of childhood atopic dermatitis at a population scale. The carrier frequency of FLG variants in the background population was reported as between 8.8% and 9.6% [7, 25, 26]. Marenholz et al. estimated that a sizeable proportion of 11.1% of eczema cases in the German population are attributable to null mutations in the filaggrin gene [27]. Palmer et al. found that these variants are absent in people of Asian and African descent, in whom other variants of filaggrin may exist [7]. This indicates that about 90% of eczema cases remain to be explained by other, undefined genetic factors that modify filaggrin protein processing or unknown mutations of other independent genes involved in the skin barrier function.
Recent GWASs of European, Chinese and Japanese populations for atopic dermatitis and a meta-analysis of GWASs have identified six new susceptibility loci at a genome-wide level of significance and among them a common variant on chromosome 11q13.5 (rs7927894[T]) [10, 28, 29]. The association of this variant with atopic dermatitis has been reported in two independent case-control study populations from Ireland and Austria and in the population-based ALSPAC (Avon Longitudinal Study of Parents and Children) birth cohort [11, 12, 30]. In contrast, a case-control study in the Japanese population did not reproduce these results [13]. A recent study found a slight association between the rs7927894[T] allele and atopic dermatitis, identifying this variant as a suggestive locus in the Chinese Han population [31]. In our study we have confirmed the association of the T allele of rs7927894 with atopic dermatitis in a population of Polish children, providing replication evidence for this genetic association. However, the effect size was a little higher than in the discovery cohorts. In our analysis the homozygous carriers of risk allele showed an OR of 2.71 (95% CI: 1.09–6.71; p = 0.028) compared to an OR of 1.46 (95% CI: 1.29–1.68) in European adult cases [10] and an OR of 1.63 (95% CI: 1.18–2.24) in Irish paediatric cases [11]. On the other hand, the frequency of the rs7927894 risk allele (15% of homozygotes) was very similar in our control group to that reported in the European control population (approx. 13% of homozygotes) [10] and in the Irish control population (approx. 14.5% of homozygotes) [11]. This may suggest that the rs7927894[T] effect may be stronger in very young children as our case series comprised children younger than 2 years, in contrast to the European adult population studied by Esperza-Gordillo et al. [10] and to the Irish paediatric population where most cases (60%) were older than 2 years [11].
The locus rs7927894 is located in an intergenic region 38 kb downstream of the C11orf30 region (chromosome 11 open reading frame 30) and 68 kb upstream of LRRC32 (leucine rich repeat containing 32). RT-PCR analysis showed that both C11orf30 and LRRC32 are potential candidates for atopic dermatitis as they are ubiquitously expressed in relevant tissues, such as skin and peripheral blood mononuclear cells [10]. The C11orf30 region encodes the nuclear protein EMSY, which binds and inactivates the cancer susceptibility gene BRCA2, and might therefore play a role in the epithelial barrier function. An increase in C11orf30 copy number has been reported in epithelium-derived cancer of the breast and ovary [10, 32]. The C11orf30 region is also associated with Crohn’s disease and inflammatory bowel disease [33]. These findings strongly suggest a potential role for C11orf30 in epithelial immunity, growth, and differentiation. The LRRC32 gene is also a plausible, or even more likely, causal candidate in this region as it has been shown that LRRC32 (also known as glycoprotein A repetitions predominant, GARP) is expressed on the surface of activated human regulatory T cells (Treg), regulates the bioavailability of transforming growth factor-β (TGF-β) and plays a crucial role in the modulation of peripheral tolerance and T effector cell function [34–37]. Additionally, Li et al. reported that four SNPs in the C11orf30-LRRC32 region were strongly associated with total serum IgE levels, concluding that this region may represent a common locus for atopic diseases through biological pathways involved in the regulation of total serum IgE levels [38].
Interestingly, our study revealed an equally strong effect of rs7927894[T] in patients with allergic and non-allergic atopic dermatitis, demonstrating that the development of atopic dermatitis in rs7927894[T] carriers is not dependent on allergic sensitisation. In contrast, a recent study in the Chinese Han population found that the rs7927894[T] allele was significantly related only to atopic dermatitis with a family history of atopy [31]. Marenholz et al. also reported that in the ALSPAC cohort there was a stronger effect of the risk variant on the atopic form of eczema and only a marginal effect in the absence of atopy. However, this study clearly demonstrated that atopy per se was not associated with the risk allele, indicating that atopy is not the primary disease manifestation of the risk variant on chromosome 11q13.5 [12].
Unexpectedly, we are not able to find significant associations between atopic dermatitis severity and either the rs7927894 SNP or the combined FLG variants. An explanation for the lack of these associations could be the study design using a general population. Previous studies showing a positive association between FLG mutations and atopic dermatitis have focused on moderate-or-severe atopic dermatitis cases recruited from specialist clinics and on children with atopic dermatitis as part of atopy-related birth cohort studies [7, 25–27]. It is possible that an association existed in Polish children with moderate-or-severe atopic dermatitis but unfortunately most of our participants have mild (76.7%) or moderate (23.3%) atopic dermatitis. On the other hand, few studies have reported the lack of association between FLG mutations and severity of atopic dermatitis [39, 40]. In relation to rs7927894 SNP, only one study evaluated a correlation with atopic dermatitis severity showing the lack of this association [31]. Our results are in agreement with this study suggesting that the severity of atopic dermatitis may not be influenced by rs7927894 SNP part. However, it cannot be excluded that the lack of this association can be observed for the same reasons that were mentioned in relation to FLG mutations.
Finally, we examined the possibility of the combined effect of rs7927894[T] and FLG loss-of-function mutations as in the case of multifactorial disease the predisposition might be better explained by a combination of alleles than just one allele. An evaluation of the distributions of rs7927894 and combined FLG null genotypes in atopic dermatitis subjects raises the possibility of gene-gene interaction. FLG loss-of-function mutations and rs7927894[T] were associated separately with an increased risk of atopic dermatitis. However, the highest risk was observed in subjects who combined both genetic risk factors. This might suggest some possible synergistic effect of the tested variants on atopic dermatitis, because the combined disease risk was higher than that observed for these alleles separately. It is possible that an involvement of rs7927894[T] and FLG mutations in a common functional pathway linked to the barrier defect is the underlying cause of the apparent interactive effect. Our results are compatible with previous findings in the ALSPAC and MAS (The German Multicentre Atopy Study) birth cohort studies that have reported the synergistic effect of rs7927894[T] and FLG mutations on the risk of eczema [12].
A potential limitation of our study, typical of all case-control studies, is a relatively small sample size and, as a consequence, rather low statistical power, which may lead to false negative or false positive results. Nevertheless, we were able to demonstrate statistically significant and biologically plausible effects. An interaction analysis requires a large population so our results should be interpreted carefully, there were some evidence on an interactions on additive scale, but there was insufficient statistical power to either confirm of refute these findings. However, they are in agreement with the outcomes of large population-based birth cohort studies that lend credibility to our results.
Conclusions
Our results indicate that the rs7927894 variant on chromosome 11q13.5 may play a role in the development of atopic dermatitis. We have provided independent replication of the association of rs7927894[T] with atopic dermatitis in the population of Polish children, however this effect seems to be independent of allergic sensitisation. To the best of our knowledge, this is the first study of the risk variant on chromosome 11q13 in this population. In addition, our results revealed that the rs7927894[T] variant and FLG loss-of-function mutations are candidate genes that might control the risk of atopic dermatitis in an interactive manner. Functional studies will be required to investigate the exact biological mechanism of these gene-gene interactions. We have concluded that future studies on atopic dermatitis have to consider gene-gene interaction as each genetic variant may have its own effect in isolation but the combination of functional SNPs in more than one gene may magnify their impact on the disease.