Purpose
Non-melanoma skin cancer (NMSC) is the most common type of skin cancer [1,2,3,4,5]. Therapeutic options such as cryotherapy, laser therapy, topical treatment, and photodynamic therapy are usually reserved for an early stage, low-risk, and superficial NMSC, while surgery and radiotherapy remain the mainstays of treatment in high-risk NMSC [6,7,8,9,10,11,12,13,14]. Skin radiotherapy can be delivered by superficial/orthovoltage X-rays, electron beams, megavoltage photons, or brachytherapy (also known as contact radiotherapy and interstitial interventional radiotherapy, BT) [7]. Unfortunately, despite sufficient data in literature, the effects of radiotherapy in NMSC are not evident, as there is only one important publication – an old randomized prospective trial [15,16].
In the past years, external beam radiotherapy (EBRT) and BT procedures became more frequent and are performed with up-to-date technology. Nowadays, multiple imaging modalities are available, which can support target volume definition and may lead to an adequate treatment planning tailoring of volumes in order to spare normal tissues as much as possible [17,18,19,20,21,22]. Differences in optimal treatments between individual patients are not only observed in case of systemic therapies but are also reported in radiotherapy [23,24,25,26]. The concept of “modern personalized medicine” redefines what is currently regarded as “the standard treatment” for each patient. It means that the combination of surgery and radiotherapy could be the right treatment option regarding function and cosmesis preservation. However, new diagnostic tools such as genomic and proteomic profiling as well as anatomical and functional imaging techniques may be used to develop predictive models for optimal clinical decision making [27,28,29,30]. It is well known that our cognitive capacity can integrate only few factors in order to make a decision, but the variables that the physicians need to consider are exponentially growing and will only increase in the future [23]. To overcome this problem, large databases can be defined to improve the quality and efficiency of treatment. Furthermore, a large database can provide the availability of significant number of patient data, and the subsequent analysis can deliver the results faster than through clinical trials. However, in order to adequately collaborate, it is necessary to ensure that all specialists fully understand the requests and the information provided. It is important to create a formal representation of a specific situation (in this case related to the domain of patients affected by skin cancer) as perceived and organized by a representative and using common language – “ontology”. An ontology is a set of concepts and categories in a domain of subjects, which shows their properties and their inter-relations. Through ontological characterization of the information, it is possible to find, isolate, organize, and integrate its meaning [31,32,33,34].
With this approach, it is expected that clinical research on skin cancer will be characterized by a less confusing understanding of considered variables, without differences in storage and interpretation. Moreover, a large database can also increase the number of variables that can be collected. These variables include clinical, therapeutic, and technical developments. The quality of collected data can be significantly improved by standardized data collection (SDC) approach, through a classification of variables that should be collected, and through regulating the most appropriate ways to measure them.
In addition to other experiences [35,36,37,38], the SKIN-COBRA (Consortium for Brachytherapy data Analysis) project was discussed and developed in the frame of the Head & Neck and Skin working group of Groupe Européen de Curiethérapie – European Society for Radiotherapy & Oncology (GEC-ESTRO). The primary model is based on patient variables (e.g., age, sex), clinical presentations of the disease (e.g., staging, markers, imaging data), treatment data (e.g., BT, chemotherapy, target therapy, immunotherapy, EBRT, surgery information, palliative care), and imaging data (diagnostic, treatment, or follow-up images). The primary objective of the SKIN-COBRA ontology is to define a specific terminological system for standardized data collection of skin cancer patients treated with BT.
Material and methods
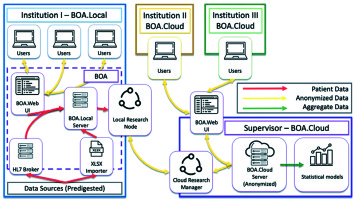
The structure of the project, the consortium agreement text, and the minimal requirements for each aspiring center were discussed and defined in the frame of the Head & Neck and Skin GEC-ESTRO Working Group (WG). The group also used a GANTT chart to schedule project workload and deadlines [39]. The centers that joined the consortium signed an agreement. The ontology was defined by a specific task group in the frame of the consortium (VL, LT, JLG, AR, GK), and KBO-laboratories (knowledge-based oncology laboratories) technical commission (KBO-TeCo) including a mathematician (AD), a PhD big data engineer (CM), a physicist (JL), a physician expert in big data, a radiomic analysis (ND), and a software expert (NDC). This multidisciplinary team defined characteristics to accept the ontology including the definition of data type for each field, the values allowed, the cardinality of items (e.g., single-select or multi-select fields), the range of numerical values, and taxonomic semantics. All these requirements aim to define the knowledge that characterizes the physicians’ specialized lexicon as precisely as possible (http://www.openclinical.org/ontologies.html). The ontology was discussed and improved by an independent multidisciplinary external committee (radiation oncologist – BF, BG, AC, MA, SR, AD, VV; dermatologist – ADS; pathologist – FF; medical oncologist – ER; plastic surgeon – SG; geriatric oncologist – GC). As a final step, the ontology was discussed and approved by the consortium. Later, the consortium defined the tools needed to share the ontology among all involved centers in a standardized way. The selected software for this approach was BOA (Beyond Ontology Awareness) (Figure 1). This software is capable of reproducing the structure of ontology, manage the legacy data import wherever needed, and coordinate data sharing activities.
All the centers within the consortium are required to:
1A. Install the service handling the ontology-compliant case report forms (CRFs) on their servers and login into the service through a web browser.
OR
1B. Connect to the centralized repository and manage the CRFs saved on their database directly through a web browser.
2. Upload legacy data into the software’s database.
3. Manually input non-legacy or new data into the desired CRFs.
4. If instruction 1A was chosen, test the connection to the centralized repository to enable data sharing.
5. Share the data according to the agreed procedure.
6. Upload legacy data into the software’s database.
7. Test the connection to the centralized repository for data sharing.
8. Share the data according to the agreed procedure.
Results
Eight centers signed the SKIN-COBRA consortium agreement (Table 1). The Skin Ontology task group, defined by the consortium committee, prepared the above described ontology table proposal (Table 2). After the characteristics of the ontology were defined, an Excel (Microsoft) template was released containing all the properties that each field had to be associated with. These properties included item name, description label, units of measurement when applicable, item number, response option text when applicable (values of tabular fields), response option value (identifiers of response option text), cardinality of response options (single-select or multi-select fields), data type, validation pattern, a flag to state if the field is a required one. There were 290 of defined variables. Each variable presented 7 properties: description label, left item text, units, response type, response options text, response values or calculations, data type. The variables are organized in 10 forms (Table 3) as follows:
Table 1
SKIN-COBRA consortium requirements
Table 2
SKIN-COBRA framework
Table 3
Forms of SKIN-COBRA ontology
1. Registry and history; 2. Histology; 3. Staging; 4. Surgery; 5. External beam radiotherapy (radical, adjuvant); 6. Systemic treatment (neoadjuvant, concomitant, adjuvant); 7. Brachytherapy (radical, adjuvant); 8. Follow-up; 9. Outcome; 10. Images and treatment files. Field types were defined as: text, number, date, table, files. DICOM (digital imaging and communication in medicine) and .txt file formats were chosen respectively for images and data treatment. The toxicity data was evaluated according to the common toxicity criteria adverse effect (CTCAE) v. 4.0. and the Radiation Therapy Oncology Group/EORTC acute and late toxicity scale (RTOG/EORTC). The RTOG/EORTC scale of choice was the preferred one as a great amount of data was stored using this scale, and direct mapping with CTCAE v. 4.0 was not possible. Data were gathered within three main levels: 1. The registry level included the minimal required information (age, gender, ethnicity, etc.) and used for epidemiological analysis only; 2. The procedure level involved treatment information and oncological outcomes; 3. The research level included the evaluation of diagnostic, treatment, and follow-up imaging available for advanced researches in individualized medicine purposes. Imaging features were developed by several studies such as radiomics analysis linking to image information with treatment and/or toxicity outcomes or gene expressions. Radiotherapy and imaging information could be automatically extracted from treatment planning system (TPS), picture archiving, and communication system (PACS), with no extra burden for data managers and figures involved. The ontology was available as a supplementary material (ontology.xls).
COBRA architecture characteristic
BOA was chosen in order to implement and propagate the ontology, as the IT architecture of BOA not only facilitates the conversion of legacy pathology archives according to the global data dictionary, but if desired, anonymously processes the clinical data based on two distinct approaches: local and centralized processing of available data (Figure 1).
The anonymized cloud-based large database was the only asset that could be shared among consortium members; this procedure was only temporary, research bounded, lasting through a lifespan of particular study. The system guarantees that nothing, except anonymous and non-referable clinical data (with no link to the original local archives) was going to become a part of the large database.
It must also be noted that while generating a predictive model by using information from one or more institutes, all statistical algorithms would only process the aggregate data, without a need to give other institutes access to individual patient records.
Discussion
The results of this project demonstrated that it is possible to create a “common language” (ontology) that connects all participating centers and, through the legacy data import procedure, information can be easily collected, allowing to take part in larger research projects, even with legacy data. Data collected in every single center is translated into the predefined ontology, which then allows to easily select groups of patients and/or groups of covariates to be anonymized, encrypted, and sent to the shared repository. The authors believe that this ontology is a viable solution to a multi-dimensional logistic dilemma involving data collection, retrieval, and applicability.
Heterogeneity is a major confounder when interpreting data from skin cancer studies, and two recent NMSC meta-analyses have presented this matter [35,36]. Both studies, despite having the opportunity to evaluate the data from thousands of patients, recommended conducting prospective trials. SKIN-COBRA has the potential to address this issue.
The challenge, which we aim to overcome through large databases, is to develop multi-factorial predictive models (using historical data to predict future events) to create a decision support system (DSS) to “personalize” the treatment both in terms of efficacy and toxicity [37,38,40,41].
The SKIN-COBRA has some unique features. Our system is absolutely in line with the EU General Data Protection Regulation (GDPR); it may act as a broker, a third-party acting as a connection between the centers participating in a cloud computing service and facilitating the distribution of data among the centers. It may also be used for implementing distributed learning and can be very helpful for privacy preservation.
The present work proves that the definition of an ontology can be used for COBRA as well as for other types of databases. Therefore, the clinician who wants to carry out researchers finds the dataset already configured, with the potential of further customization.
Large databases in radiation oncology mean studying large cohorts of patients and integrating heterogeneous types of data. The science of oncology is moving away from standardized therapies, in which decisions are based only on anatomical, histological, and molecular features that define groups of patients and diseases to personalized approach. This evolution implies a large amount of available data, which will influence physicians in their clinical practice. Furthermore, it allows to choose and combine the best and most up-to-date treatment evidence in a tailored approach. The use of large database is the simplest way to digitally capture a large amount of data regarding patient characteristics, treatment features, adverse events, and follow-ups. It can be used to generate new knowledge, which may help physicians in this task, providing predictive models and decision support tools (DSS).
By using this DSS it is possible to analyze and interpret the available data to formulate hypotheses and to obtain useful answers for early diagnosis and better treatment. Focusing on the characteristics of individual patient permit to follow the effectiveness, efficiency, and correctness that are typical for current medicine as well as the use of technologies capable of refining and innovating the classic epidemiological methodologies on vast amounts of data [27,28,29]. Promoting and supporting a predictive, preventive, and personalized medicine also means dealing with the costs of healthcare. The data must be of qualitative validity, producing credible results that can guide the specialist to make a decision based on the correlations discovered by the model, and to use this additional information to integrate with already possessed knowledge. The DSS should simultaneously predict local control, survival, treatment toxicity, quality of life, and cost. This system should represent these predictions and stabilize the information to be easily understood for the physicians as well as for the patients, allowing shared decision making. Large database analysis results have the potential to guide decisions at the time of initial consultation for the best treatment options, according to the individual patient’s feature, and state of knowledge. The kind of radiotherapy, treatment time, associated chemotherapy, targeted therapy, or immunotherapy would be chosen not only by the physician, but by an algorithm supporting the role and experience of physicians.
The same system could also guide treatment decisions for eventual adverse event management. Additionally, it is useful after the treatment for follow-up and early detection of a relapse. In the future, follow-up needs to integrate all the data collected by wearable devices and connected objects that are being adopted by a large proportion of the population [28]. Continuous and real-time monitoring of abnormal events would lead to earlier detection of relapse and optimization of salvage treatment’s efficiency and cost. Eventually, overall survival will be impacted by such approaches [29].
The combinations of the various parameters can also be represented with nomograms, typical tools that allow the assignment of scores and graphic scales display of the variables that have prognostic relevance.
Radiotherapy is one of the main protagonists of this development, and the number of in-use large databases is increasing fundamentally on the basis of three important phenomena.
The first phenomenon is represented by the digital improvement of diagnostic imaging, which attempts to prevent and early diagnose of the disease; the emergence of new powerful digital diagnostic technologies is visible. The diffusion of these tools has led to the creation of DICOM standard, which defines the guidelines for storing and sharing images. COBRA allows an internal or external validation of the predictive models, so it could provide decision support tools classified also with maximum level of TRIPOD classification [42].
The second phenomenon is connected to the folders and electronic files of patients, since paper files have been eliminated in many centers for a decade or more. The goal is to achieve significant improvements in patient care and not to limit oneself to a mere adoption and compilation of electronic files.
The last phenomenon is characterized by the development of biotechnologies used in the field of the so-called “omics” sciences (genomics, transcriptomics, proteomics, metabolomics, epigenomics, radiomics, etc.). Here, the goal is to provide personalized therapies, which are aimed at the characteristics of individual and inspired by personalized medicine.
Conclusions
Large databases are a natural extension of traditional statistical approaches, a valuable and increasingly necessary tool for a modern healthcare system. The future analysis of the collected multinational and multi-institutional data will show if the use of the system can result in high-quality evidence in the multidisciplinary treatment of NMSC, thus helping to use this information for personalized treatment decisions.